How to Upload Internal Data When Creating an R Package Sysdata.rda
There are already a lot of great resources1 that teach you how to make an R package. Simply I thought it would still be worthwhile to walk through how I created my outset i – at the very least, it'll be helpful for myself when I write my adjacent R bundle.
Here is my step-by-step guide to building a (minimally functional) R package.
Pace 1: Set up your packages and directory
To create your ain R package, you will demand to employ 2 packages, devtools and roxygen2. devtools contains the functions for edifice a package, while roxygen2 helps generate documentation for the parcel using specially formatted comments.
library(devtools) library(roxygen2) At present navigate to the parent directory of where you desire your R package directory to be located (east.thou.R_packages, but information technology can be wherever you want it to exist). Utilize the function devtools::create 2 to create the binder for the R package, which I will call mypackage in this tutorial.
setwd("R_packages") create("mypackage") Your mypackage directory volition at present exist under your parent directory. It should look similar the following:
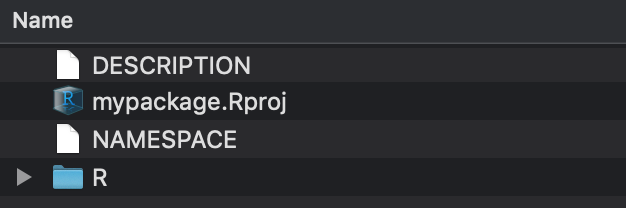
Contents of mypackage:
-
Clarification- information about the parcel -
mypackage.Rproj- the R project file -
NAMESPACE- data about what to import and export -
R- a directory for your scripts, i.e. where your functions will go
At this point, I recommend that you make your R packet a git repository and connect it to GitHub. To do so, navigate to https://github.com and create a new repository there with the aforementioned name (e.m. "mypackage"). And so run the following commands in your final.
cd mypackage # Navigate to mypackage directory git init git remote add together origin https://github.com/albertkuo/mypackage.git git add * git commit -k "Initial commit" git push -u origin master Aside from the usual benefits that come with using version command, making your R bundle a GitHub repo ways that afterward, other people can easily install your package directly from GitHub (see Step vi).
Step 2: Write Clarification
Now open mypackage.Rproj in RStudio.3 Let's edit the Description file starting time. You can practise this in RStudio or with your favorite text editor. The Clarification file basically contains some information about the parcel. Below is an example (you will take to remove the "# comments" in your bodily DESCRIPTION file).
Package: mypackage Title: What the Packet Does # One line only, Title Instance, and no catamenia Version: 0.ane.0 # see Jeff Leek's versioning convention Date: 2019-12-14 Authors@R: c(person(given = "Albert", family = "Kuo", role = c("aut", "cre"), # "aut" ways author (someone who has # made a significant contribution) # "cre" ways maintainer (who to # contact if in that location are problems) email = "albertkuo@jhu.edu", comment = c("ORCID" = "0000-0001-5155-0748"))) Depends: R (>= three.6.0) Imports: dplyr # packages you need for the bundle, aka dependencies Suggests: knitr # optional packages that aren't strictly needed Clarification: What the package does, with more particular. # ane paragraph License: GPL-3 Encoding: UTF-8 LazyData: truthful Note: The GPL-3 license seems to be a commonly recommended license for R packages. I found this guide to be a helpful start if you lot want to learn more than well-nigh software licenses.
Pace 3: Add functions and write documentation
It's likely that you already accept functions that y'all want to create a package for. Copy them into the R folder. Each function should be in its own split file.4
Let'southward say yous have a script called my_function.R. We want to write peculiarly formatted comments for it, i.e. the #' comments, so that roxygen2 tin can automatically build the documentation pages. Beneath is an instance.
#' A very brief clarification of what your function does #' #' A detailed description of what the function does. #' #' @param print_statement a logical value indicating #' whether to impress the argument (default is \code{TRUE}) #' #' @import dplyr #' #' @export #' #' @return \lawmaking{my_function} prints a declaration #' announcing itself if the parameter is \lawmaking{TRUE} #' #' @examples #' R code examples of how to apply your function #' my_function <- role(print_statement = True){ if(print_statement){ print("This is my function.") } } Some things I want to draw your attention to:
- Include
@exportif you lot want the function to be exported, i.e. visible to people who apply the R bundle. - Include
@importfor any packages that are needed for this part. Y'all tin likewise import a specific office with@importFromto prevent name conflicts (e.g.@importFrom MASS lda).
Now we can generate the documentation files and NAMESPACE using roxygen2.five This is equally easy as typing the following command.
document() Your mypackage directory should now look like the following:
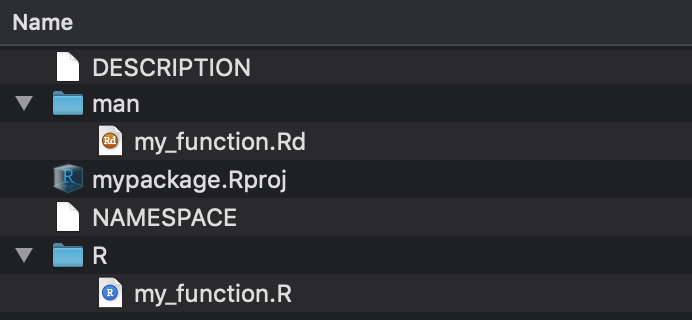
New contents of mypackage:
- In the
mandirectory are the documentation files. These are the help files yous encounter when yous blazon?my_function. - The NAMESPACE file tells yous which functions you lot are exporting from your package and which packages/functions yous are importing to exist used in your package.
Every time you make changes to the comments, you volition want to regenerate the documentation and NAMESPACE with the devtools::document() control.
(Optional) Footstep iv: Add internal data
Something that you might desire to practice is to employ data internally within your R package. For example, your functions might require the utilise of some internal data that you want to load.
To do this, get-go run the following command.
use_data_raw("generate_internal") This will create an R script in data-raw/generate_internal.R, where yous can put your code that generates the internal data. It will also add the data-raw/ directory to .Rbuildignore, since y'all won't need it to build the parcel.
And then in generate_internal.R, you tin can generate and save the internal data.
# Generate data x = 1:5 y = 1:ten # Save information usethis::use_data(x, y, internal = Truthful) The command usethis::use_data will save the objects x and y in R/sysdata.rda, which is a file specifically designed to hold internal information. Your functions will then be able to phone call the objects x and y whenever they are needed.
Pace five: Install your package
Believe it or not, your parcel is prepare to exist installed locally on your computer. This is uncomplicated to do with the devtools::install() command.
install() library(mypackage) Your package is at present fix to be used! Whatever functions you lot've exported are bachelor. For instance, blazon my_function(). You can also access the assist page you lot generated in Pace 3, just like you lot would exercise with whatever other package'southward functions, past typing ?my_function.
Futurity work
At this signal, you have a basic R bundle that can be hands distributed. Fifty-fifty if you don't share it with anyone, this is a slap-up fashion to organize and utilize your own functions.6
To plow your R parcel into something more user-friendly, however, there are a couple more things you lot will desire to practice. One of them is to write tests for your R package to cheque for bugs and to "dummy proof" your functions. Another is to write vignettes to help people learn how to use your R package. Since these things go beyond what is strictly necessary for a functional R package,7 I will write well-nigh them in a later on postal service.
Source: https://blog.albertkuo.me/post/creating-an-r-package-for-the-first-time/
0 Response to "How to Upload Internal Data When Creating an R Package Sysdata.rda"
Post a Comment